About
This project will yield three main deliverables:
1) Contributions to GA4GH APIs and toolkits for software infrastructure, data security and ethics, to support epigenomic data; 2) Development of a platform, including a portal, for epigenomic data and metadata, using GA4GH tools from the first deliverable; and 3) Applications that leverage this infrastructure to answer user needs in real projects handling epigenomic data.
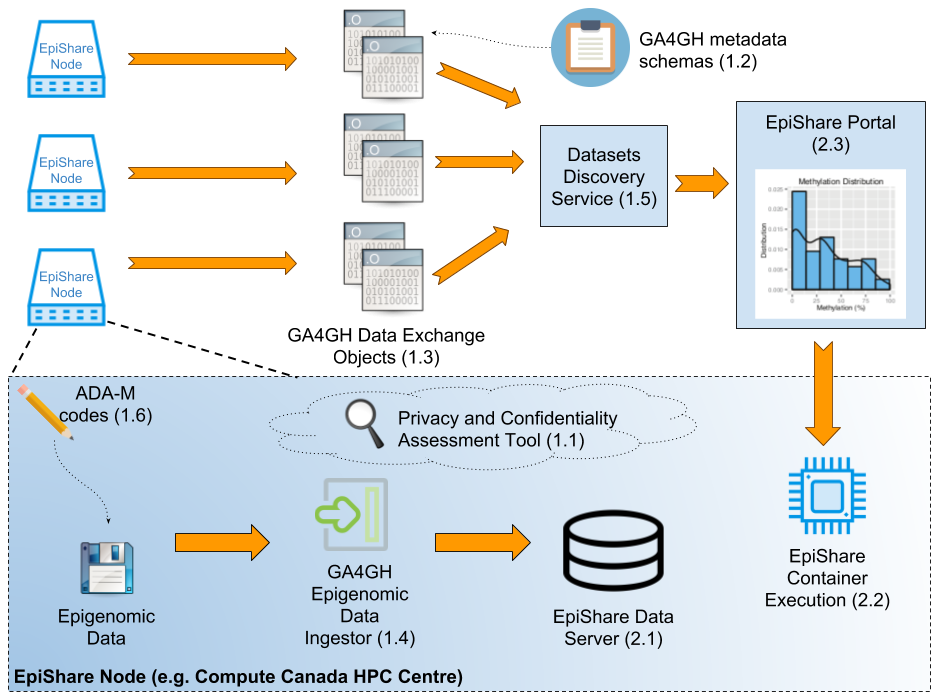
EpiShare Nodes are the repositories to epigenomic data, deployed at sites such as Compute Canada HPC centres. The Nodes data privacy considerations are assessed using Privacy and Confidentiality Assessment Tool (1.1). When adding new datasets to a Node, consent and access conditions to which the data is bound are made available as queryable metadata using GA4GH ADA-M (1.6). Datasets are then processed by the GA4GH Epigenomic Data Ingestor (1.4) to generate searchable metadata on epigenomic features (e.g. Methylation ratio at CpG islands, ChIP-Seq peaks location).
Data is then stored on the EpiShare Data Server (2.1). Each EpiShare Node shares metadata on available datasets with the Discovery Service (1.5) using GA4GH Data Exchange Object (1.3), following GA4GH metadata specifications (1.2). The EpiShare Portal (2.3) is able to search these datasets based on metadata, or epigenetic features (e.g. I’d like Bisulfite-Seq datasets where Methylation Ratio is > 50% at a given genomic position). The Portal can then launch an Analysis Container (2.2) directly on the EpiShare Node to analyse selected datasets. Analysis results will be returned to the EpiShare Portal.
Sponsors

